Single-cell transcriptomics identifies chondrocyte differentiation dynamics in vivo and in vitro
Developmental Cell (2025)
Lawrence JEG, Woods S, Roberts K, Sumanaweera D, Balogh P, Li T, Predeus AV, He P, Polanski K, Prigmore E, Tuck E, Mamanova L, Zhou D, Webb S, Jardine L, He X, Barker RA, Haniffa M, Flanagan AM, Young MJ, Behjati S, Bayraktar O, Kimber SJ, Teichmann SA
A novel human fetal lung-derived alveolar organoid model reveals mechanisms of surfactant protein C maturation relevant to interstitial lung disease
EMBO J (2025)
Lim K, Rutherford EN, Delpiano L, He P, Lin W, Sun D, Van den Boomen DJH, Edgar JR, Bang JH, Predeus A, Teichmann SA, Marioni JC, Matesic LE, Lee JH, Lehner PJ, Marciniak SJ, Rawlins EL, Dickens JA
Considerations for building and using integrated single-cell atlases
Nat Methods (2024)
Hrovatin K, Sikkema L, Shitov VA, Heimberg G, Shulman M, Oliver AJ, Mueller MF, Ibarra IL, Wang H, Ramírez-Suástegui C, He P, Schaar AC, Teichmann SA, Theis FJ, Luecken MD
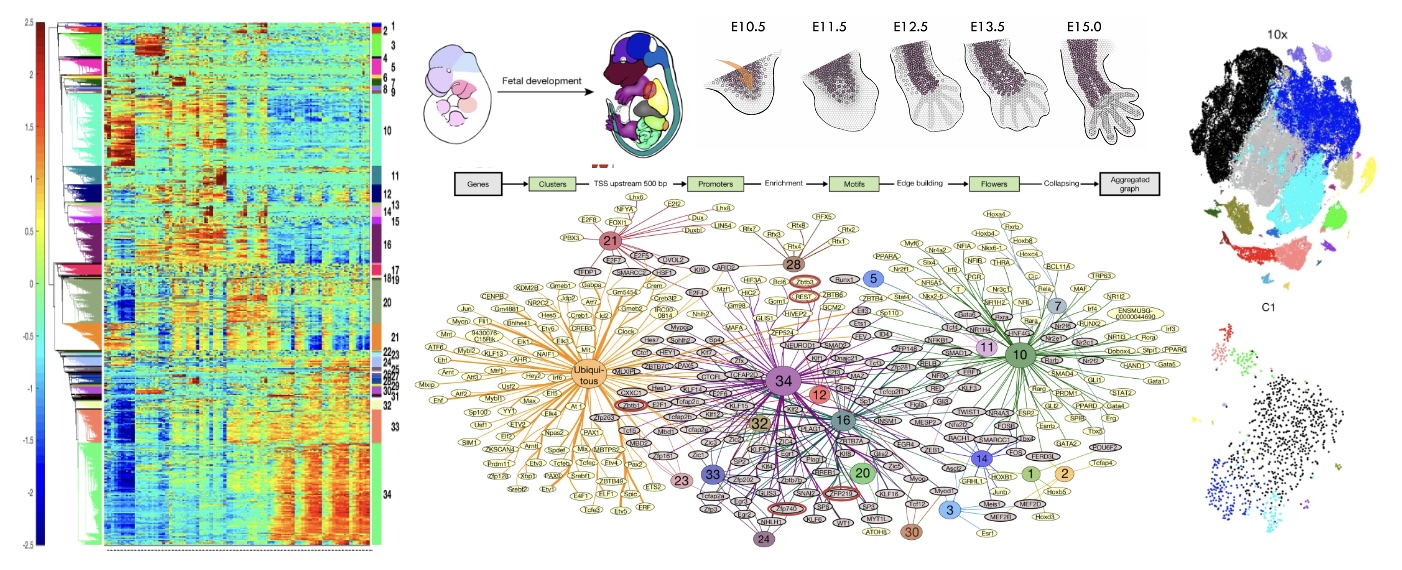
A multi-omic atlas of human embryonic skeletal development
Nature (2024)
To K, Fei L, Pett JP, Roberts K, Blain R, Polanski K, Li T, Yayon N, He P, Xu C, Cranley J, Moy M, Li R, Kanemaru K, Huang N, Megas S, Richardson L, Kapuge R, Perera S, Tuck E, Wilbrey-Clark A, Mulas I, Memi F, Cakir B, Predeus AV, Horsfall D, Murray S, Prete M, Mazin P, He X, Meyer KB, Haniffa M, Barker RA, Bayraktar O, Chédotal A, Buckley CD, Teichmann SA
A prenatal skin atlas reveals immune regulation of human skin morphogenesis
Nature (2024)
Gopee NH, Winheim E, Olabi B, Admane C, Foster AR, Huang N, Botting RA, Torabi F, Sumanaweera D, Le AP, Kim J, Verger L, Stephenson E, Adão D, Ganier C, Gim KY, Serdy SA, Deakin C, Goh I, Steele L, Annusver K, Miah MU, Tun WM, Moghimi P, Kwakwa KA, Li T, Basurto Lozada D, Rumney B, Tudor CL, Roberts K, Chipampe NJ, Sidhpura K, Englebert J, Jardine L, Reynolds G, Rose A, Rowe V, Pritchard S, Mulas I, Fletcher J, Popescu DM, Poyner E, Dubois A, Guy A, Filby A, Lisgo S, Barker RA, Glass IA, Park JE, Vento-Tormo R, Nikolova MT, He P, Lawrence JEG, Moore J, Ballereau S, Hale CB, Shanmugiah V, Horsfall D, Rajan N, McGrath JA, O'Toole EA, Treutlein B, Bayraktar O, Kasper M, Progatzky F, Mazin P, Lee J, Gambardella L, Koehler KR, Teichmann SA, Haniffa M
WebAtlas pipeline for integrated single-cell and spatial transcriptomic data
Nat Methods (2024)
Li T, Horsfall D, Basurto-Lozada D, Roberts K, Prete M, Lawrence JEG, He P, Tuck E, Moore J, Yoldas AK, Babalola K, Hartley M, Ghazanfar S, Teichmann SA, Haniffa M, Bayraktar OA
A single-cell atlas enables mapping of homeostatic cellular shifts in the adult human breast
Nat Genet (2024)
Reed AD, Pensa S, Steif A, Stenning J, Kunz DJ, Porter LJ, Hua K, He P, Twigger AJ, Siu AJQ, Kania K, Barrow-McGee R, Goulding I, Gomm JJ, Speirs V, Jones JL, Marioni JC, Khaled WT
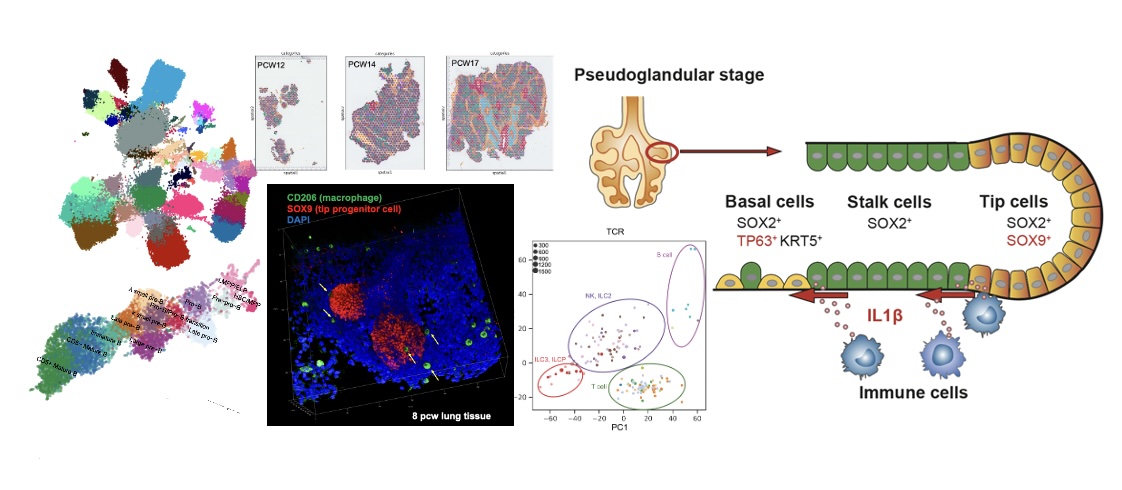
Early human lung immune cell development and its role in epithelial cell fate
Sci Immunol (2023)
Barnes JL*, Yoshida M*, He P*, Worlock KB, Lindeboom RGH, Suo C, Pett JP, Wilbrey-Clark A, Dann E, Mamanova L, Richardson L, Polanski K, Pennycuick A, Allen-Hyttinen J, Herczeg IT, Arzili R, Hynds RE, Teixeira VH, Haniffa M, Lim K, Sun D, Rawlins EL, Oliver AJ, Lyons PA, Marioni JC, Ruhrberg C, Tuong ZK, Clatworthy MR, Reading JL, Janes SM, Teichmann SA, Meyer KB, Nikolic MZ
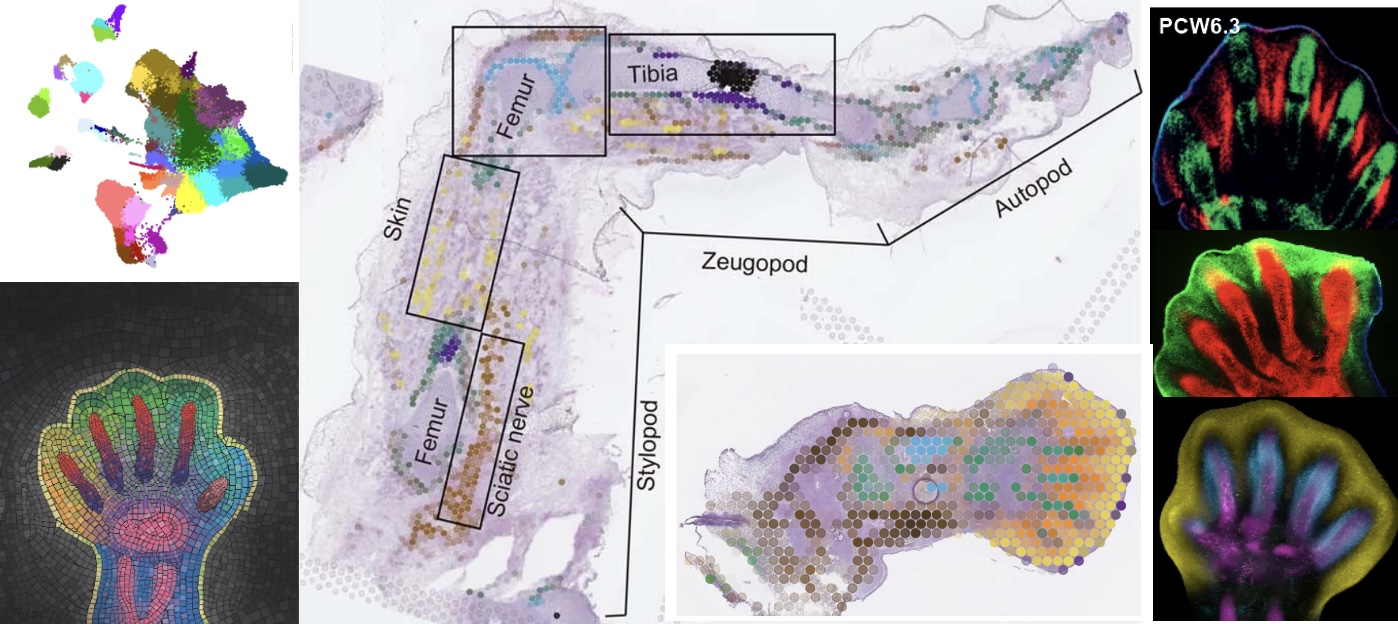
A human embryonic limb cell atlas resolved in space and time
Nature (2023)
Zhang B*, He P*, Lawrence JEG*, Wang S*, Tuck E, Williams BA, Roberts K, Kleshchevnikov V, Mamanova L, Bolt L, Polanski K, Li T, Elmentaite R, Fasouli ES, Prete M, He X, Yayon N, Fu Y, Yang H, Liang C, Zhang H, Blain R, Chedotal A, FitzPatrick DR, Firth H, Dean A, Bayraktar OA, Marioni JC, Barker RA, Storer MA, Wold BJ, Zhang H, Teichmann SA
A spatially resolved atlas of the human lung characterizes a gland-associated immune niche
Nat Genet (2023)
Madissoon E, Oliver AJ, Kleshchevnikov V, Wilbrey-Clark A, Polanski K, Richoz N, Ribeiro Orsi A, Mamanova L, Bolt L, Elmentaite R, Pett JP, Huang N, Xu C, He P, Dabrowska M, Pritchard S, Tuck L, Prigmore E, Perera S, Knights A, Oszlanczi A, Hunter A, Vieira SF, Patel M, Lindeboom RGH, Campos LS, Matsuo K, Nakayama T, Yoshida M, Worlock KB, Nikolic MZ, Georgakopoulos N, Mahbubani KT, Saeb-Parsy K, Bayraktar OA, Clatworthy MR, Stegle O, Kumasaka N, Teichmann SA, Meyer KB
Organoid modeling of human fetal lung alveolar development reveals mechanisms of cell fate patterning and neonatal respiratory disease
Cell Stem Cell (2023)
Lim K, Donovan APA, Tang W, Sun D, He P, Pett JP, Teichmann SA, Marioni JC, Meyer KB, Brand AH, Rawlins EL
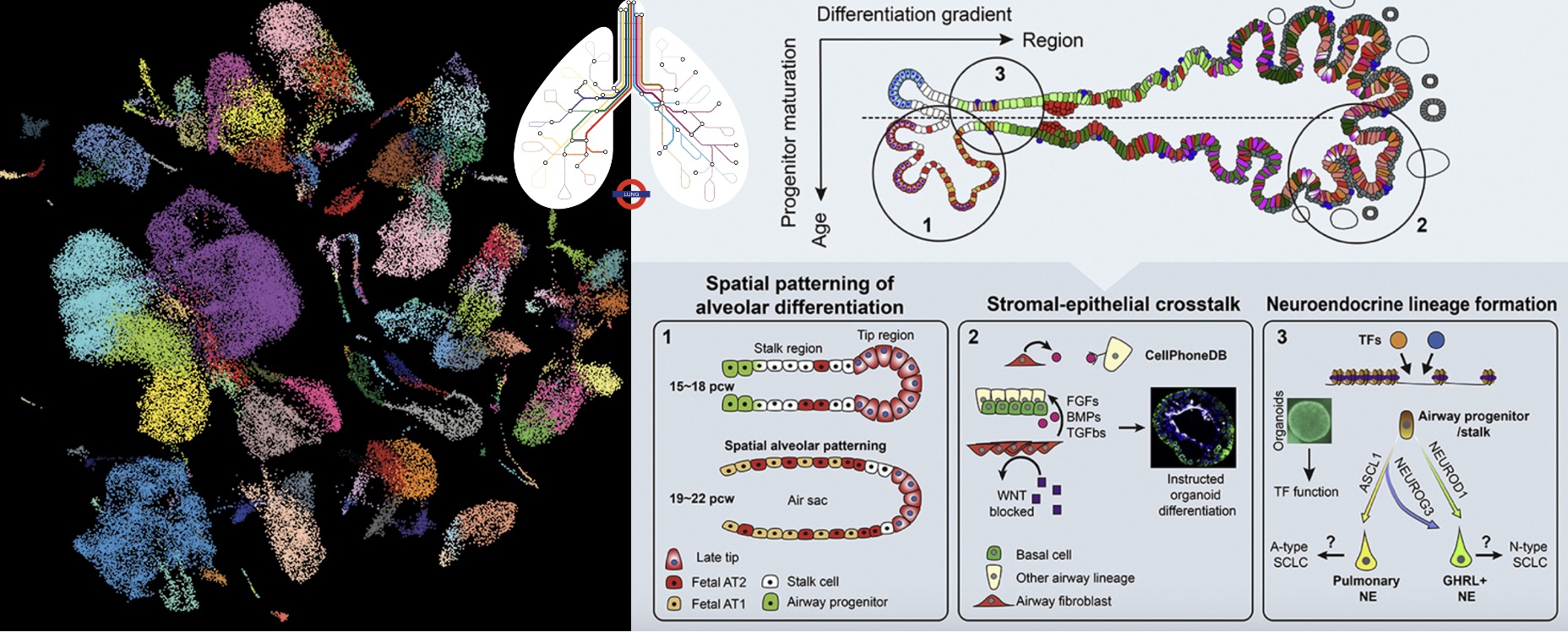
A human fetal lung cell atlas uncovers proximal-distal gradients of differentiation and key regulators of epithelial fates
Cell (2022)
He P*, Lim K*, Sun D*, Pett JP, Jeng Q, Polanski K, Dong Z, Bolt L, Richardson L, Mamanova L, Dabrowska M, Wilbrey-Clark A, Madissoon E, Tuong ZK, Dann E, Suo C, Goh I, Yoshida M, Nikolic MZ, Janes SM, He X, Barker RA, Teichmann SA, Marioni JC, Meyer KB, Rawlins EL
SOX9 maintains human foetal lung tip progenitor state by enhancing WNT and RTK signalling
EMBO J (2022)
Sun D, Llora Batlle O, van den Ameele J, Thomas JC, He P, Lim K, Tang W, Xu C, Meyer KB, Teichmann SA, Marioni JC, Jackson SP, Brand AH, Rawlins EL
Mapping the developing human immune system across organs
Science (2022)
Suo C, Dann E, Goh I, Jardine L, Kleshchevnikov V, Park JE, Botting RA, Stephenson E, Engelbert J, Tuong ZK, Polanski K, Yayon N, Xu C, Suchanek O, Elmentaite R, Domínguez Conde C, He P, Pritchard S, Miah M, Moldovan C, Steemers AS, Mazin P, Prete M, Horsfall D, Marioni JC, Clatworthy MR, Haniffa M, Teichmann SA
Single-cell meta-analysis of SARS-CoV-2 entry genes across tissues and demographics
Nat Med (2022)
Muus C, Luecken MD, Eraslan G, Sikkema L, Waghray A, Heimberg G, Kobayashi Y, Vaishnav ED, Subramanian A, Smillie C, Jagadeesh KA, Duong ET, Fiskin E, Torlai Triglia E, Ansari M, Cai P, Lin B, Buchanan J, Chen S, Shu J, Haber AL, Chung H, Montoro DT, Adams T, Aliee H, Allon SJ, Andrusivova Z, Angelidis I, Ashenberg O, Bassler K, Bécavin C, Benhar I, Bergenstråhle J, Bergenstråhle L, Bolt L, Braun E, Bui LT, Callori S, Chaffin M, Chichelnitskiy E, Chiou J, Conlon TM, Cuoco MS, Cuomo ASE, Deprez M, Duclos G, Fine D, Fischer DS, Ghazanfar S, Gillich A, Giotti B, Gould J, Guo M, Gutierrez AJ, Habermann AC, Harvey T, He P, Hou X, Hu L, Hu Y, Jaiswal A, Ji L, Jiang P, Kapellos TS, Kuo CS, Larsson L, Leney-Greene MA, Lim K, Litvinuková M, Ludwig LS, Lukassen S, Luo W, Maatz H, Madissoon E, Mamanova L, Manakongtreecheep K, Leroy S, Mayr CH, Mbano IM, McAdams AM, Nabhan AN, Nyquist SK, Penland L, Poirion OB, Poli S, Qi C, Queen R, Reichart D, Rosas I, Schupp JC, Shea CV, Shi X, Sinha R, Sit RV, Slowikowski K, Slyper M, Smith NP, Sountoulidis A, Strunz M, Sullivan TB, Sun D, Talavera-López C, Tan P, Tantivit J, Travaglini KJ, Tucker NR, Vernon KA, Wadsworth MH, Waldman J, Wang X, Xu K, Yan W, Zhao W, Ziegler CGK, NHLBI LungMap Consortium, Human Cell Atlas Lung
The changing mouse embryo transcriptome at whole tissue and single-cell resolution
Nature (2020)
He P*, Williams BA*, Trout D, Marinov GK, Amrhein H, Berghella L, Goh ST, Plajzer-Frick I, Afzal V, Pennacchio LA, Dickel DE, Visel A, Ren B, Hardison RC, Zhang Y, Wold BJ
Perspectives on ENCODE
Nature (2020)
ENCODE Project Consortium, Snyder MP, Gingeras TR, Moore JE, Weng Z, Gerstein MB, Ren B, Hardison RC, Stamatoyannopoulos JA, Graveley BR, Feingold EA, Pazin MJ, Pagan M, Gilchrist DA, Hitz BC, Cherry JM, Bernstein BE, Mendenhall EM, Zerbino DR, Frankish A, Flicek P, Myers RM
Expanded encyclopaedias of DNA elements in the human and mouse genomes
Nature (2020)
ENCODE Project Consortium, Moore JE, Purcaro MJ, Pratt HE, Epstein CB, Shoresh N, Adrian J, Kawli T, Davis CA, Dobin A, Kaul R, Halow J, Van Nostrand EL, Freese P, Gorkin DU, Shen Y, He Y, Mackiewicz M, Pauli-Behn F, Williams BA, Mortazavi A, Keller CA, Zhang XO, Elhajjajy SI, Huey J, Dickel DE, Snetkova V, Wei X, Wang X, Rivera-Mulia JC, Rozowsky J, Zhang J, Chhetri SB, Zhang J, Victorsen A, White KP, Visel A, Yeo GW, Burge CB, Lécuyer E, Gilbert DM, Dekker J, Rinn J, Mendenhall EM, Ecker JR, Kellis M, Klein RJ, Noble WS, Kundaje A, Guigó R, Farnham PJ, Cherry JM, Myers RM, Ren B, Graveley BR, Gerstein MB, Pennacchio LA, Snyder MP, Bernstein BE, Wold B, Hardison RC, Gingeras TR, Stamatoyannopoulos JA, Weng Z